
About
The generation of induced pluripotent stem cells (iPSCs), especially the generation of patient-derived pluripotent stem cells suitable for disease modelling in vitro, opens the door for the potential translation of stem-cell related studies into the clinic.
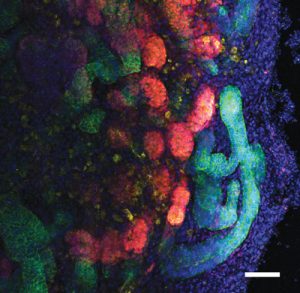
Figure 1: A. Representative immunofluorescence image of an organoid at day 25 of differentiation stained for the expression of ECADHERIN (green), WT1 (red) and PODOCALYXIN (yellow). Scale bar, 500 μm.
Successful replacement, or augmentation, of the function of damaged cells by patient derived differentiated stem cells would provide a novel cell-based therapy for diseases. Since iPSCs resemble human embryonic stem cells (hESCs) in their ability to generate cells of three germ layers, patient-specific iPSCs offer definitive solutions for the ethical and histo-incompatibility issues related to hESCs. Indeed human iPSC (hiPSC)-based autologous transplantation is heralded as the future of regenerative medicine.
One of our aims is to generate and correct disease-specific hiPSCs for disease modelling and drug screening. The combination of gene-editing based methodologies together with the development of novel protocols for cell differentiation into relevant tissues/organs, provides a unique scenario for modelling disease progression, and the identification of molecular and cellular mechanisms leading to organ regeneration (Figure 2). In this regard we are particularly interested in generation of transgene-free and disease free patient derived hiPSCs for disease modelling and the discovery of novel therapeutic targets.
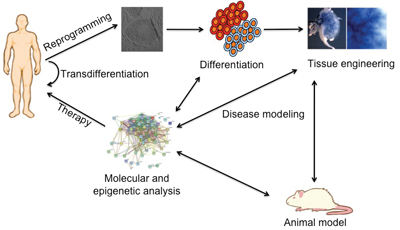
Figure 2: Patient induced pluripotent stem cells (iPSCs) represent an unprecedented tool for the generation of in vitro platforms for disease modelling and the definition of protocols for pluripotent stem cells differentiation. Transdifferentiation also offers the possibility to generate auto-compatible cells with no need to undergo to pluripotent stage. In these scenarios the correction of the genetic defect(s) leading to disease may help to understand the molecular and cellular mechanisms driving disease gestation and progression, and more importantly, to identify novel mechanisms leading to organ regeneration. The combination of gene editing methodologies with defined protocols for tissue differentiation helps us to generate in vitro systems for drug screening and disease modelling.
We believe that the recovery of tissue function should not be restricted to the development of cell replacement therapies. In this regard, in our laboratory we take advantage of organisms that possess the ability to regenerate such as zebrafish, in order to understand which molecular and cellular pathways lead to organ regeneration. Surprisingly, studies in neonatal mice have demonstrated that soon after birth this organism posses the capability to regenerate its heart. Taking advantage of such preliminary observations we are translating such analysis in order to understand if the mammalian neonatal kidney still posses the capability to regenerate, and more importantly, if we are able to dissect the epigenetic and cellular mechanisms leading to those responses.
 Lastly, and in an effort to fully develop in vitro and ex vivo platforms for organ regeneration, in our lab we are focused in the development of reporter cell lines for different transcription factors essential for tissue-specific commitment and differentiation (i.e: renal and cardiac lineages). The possibility to combine pluripotent stem cell lines together with decellularized matrices, functionalized biomaterials and ex vivo organoids offers and unprecedented opportunity for the immediate generation of patient-specific in vitro and ex vivo platforms for disease modelling and organ regeneration (Figure 3).
Lastly, and in an effort to fully develop in vitro and ex vivo platforms for organ regeneration, in our lab we are focused in the development of reporter cell lines for different transcription factors essential for tissue-specific commitment and differentiation (i.e: renal and cardiac lineages). The possibility to combine pluripotent stem cell lines together with decellularized matrices, functionalized biomaterials and ex vivo organoids offers and unprecedented opportunity for the immediate generation of patient-specific in vitro and ex vivo platforms for disease modelling and organ regeneration (Figure 3).
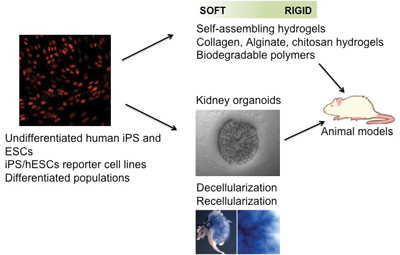
Figure 3: Induced pluripotent stem cells (iPSCs) resemble human embryonic stem cells (hESCs) in their ability to generate cells of the three germ layers of the embryo. This capacity can help us to understand the molecular and cellular cues driving cell fate. Our aim is to generate reporter cell lines from patient iPSCs in order to develop robust protocols for pluripotent stem cells differentiation. Moreover, the combination of patient differentiated populations together with functionalized biomaterials, ex vivo approaches (i.e: organoids), and decellularized tissue matrices, offers and unprecedented strategy for organ regeneration.
News/Jobs
“Las bioimpresoras abren el camino a la creacion de organos en 3D”
09/10/17
Nuria Montserrat features in an article in El Periodico today which discusses the possibilities offered by 3D bioprinting to create replacement tissues or even whole organs.
“Más cerca de generar corazones bioartificiales”
29/06/16
An article about Nuria Montserrat appeared in El Mundo on Tuesday following her invovlement in a recent study in which the first human heart grafts from human pluripotent stem cells were generated.
Researchers generate human heart grafts from human pluripotent stem cells
19/05/16
Scientists from IBEC, in collaboration with the Hospital General Universitario Gregorio Marañón in Spain and two other groups in the USA, have made a big leap in heart regeneration advances by achieving heart grafts from human pluripotent stem cells for the first time in less than one month.
Bioenginyeria per fer realitat “L’home de carn”
04/05/15
In the magazine Ara, IBEC group leader Nuria Montserrat and artist Marcel·lí Antúnez, who creates interactive sculpture with organic materials, such as Joan l’Home de Carn, appeared in an article together talking about organ regeneration.
Genetic “editing” a new tool to fight inherited disease
24/04/15
Researchers at the Hospital Clínic, IDIBAPS, the Hospital Sant Joan de Deu and the Institute for Bioengineering of Catalonia (IBEC) have participated in a study, led by Dr. Juan Carlos Izpisúa Belmonte of the Gene Expression Laboratory at California’s Salk Institute, that uses molecular “scissors” to remove mitochondrial mutations in mouse eggs.
“Miniriñones de laboratorio”
10/02/2015
New IBEC group leader Nuria Montserrat is featured in an article in El Mundo.
Projects
EU-funded projects
| REGMAMKID How to regenerate the mammalian kidney (2015-2020) | European Commission, ERC-StG | Nuria Montserrat |
National projects
| CHONDREG Identification of the epigenetic mechanisms preventing chondrocyte de-differentiation: generation of novel therapeutic strategies for the treatment of cartilage chronic osteochondral lesions | CIBER | Nuria Montserrat |
| Infarto de miocardio en jóvenes. Factores epigeneticos y nuevos marcadores de riesgo cardiovascular. Efecto de la modulación de la expresión de microRNAs y long-non coding RNAs | ISCIII | (Collaborator) |
| Desarrollo de nuevas estrategias para el tratamiento de la enfermedad renal (2015-2017) | MINECO | Nuria Montserrat |
| TRATENFREN Desarrollo de nuevas estrategias para el tratamiento de la enfermedad renal (2015-2017) | MINECO, Retos investigación: Proyectos I+D | Nuria Montserrat |
| Regenerative medicine for Fanconi anemia: generation of disease-free patient-specific iPS (2013-2016) | Fundació La Marató de TV3 | Nuria Montserrat |
Publications
Equipment
- Real Time QuantStudio 5
- SimpliAmp thermocycler
- Eppendorf 5415D centrifuge
- Allegra X-15 R centrifuge
- Gyrozen 1248 centrifuge
- BioUltra 6 Telstar culture Hood 2x
- AH-100 Telstar primary culture Hood
- Binder CB 60 incubators 2x
- Controltecnica ASTEC SCA 165 incubator
- Controltecnica ZC 180 incubator
- Bioruptor Pico sonicator
- Thermomixer C thermal block
- Leica DMS1000 and DMIL Led microscopes
- Leica DMi1 microscope
- Leica MZ 10F magnifying glass
- Safe Imager 2.0 transilluminator
Collaborations
- Juan Carlos Izpisua Belmonte
Salk Institute for Biological Studies - Dr. Josep Maria Campistol Plana
Experimental Laboratory of Nephrology and Transplantation, Hospital Clínic, Barcelona - Peter Hohestein
The Roslin Institute, University of Edinburgh - Dr. Pere Gascón Vilaplana
Head of Oncology Service/Molecular and Translational Oncology Laboratory, IDIBAPS - Gloria Calderon
Embryotools SL - Pura Muñoz Cánovas
Departament de Ciències Experimentals i de la Salut, Universitat Pompeu Fabra - Dr. Pedro Guillén
Director Clínica Cemtro, Madrid - Dr. Francisco Fernández Avilés
Head of Cardiology Service, Hospital General Universitario Gregorio Marañón, Madrid - Dr María Eugenia Fernández
Unit of Cell Production, Hospital Gregorio Marañón, Madrid - Joaquin Gutiérrez Fruitós
University of Barcelona - Dr. Elena Martínez
Biomimetic systems for cell engineering, IBEC - Dr. Cristina Eguizabal Argaiz
Centro Vasco de Transfusion y Tejidos Humanos (CVTTH), Bizkaia

