ABOUT
Our lab’s focus is on understanding the mechanisms that control mammalian embryo implantation and apply that knowledge to provide solutions that improve human assisted reproduction techniques (ART).
The embryonic development of humans (and mammals in general) requires the implantation of the embryo into the walls of the maternal uterus. This process is highly inefficient as on average, 25–30% of conceptions lead to successful live births and around 60% of all conceptions are lost at the time of (or soon after) implantation. However, despite the central role of implantation in human fertility, the process is still elusive to experimentation because of its inaccessibility.
To overcome the elusiveness of this process, the group combines imaging and bioengineering methods to efficiently culture and image pre-implantation embryos and allow them to implant outside the uterus in highly physiological conditions. Our systems are accessible to imaging tools which allow us to interrogate the genetics, metabolomics, and mechanics of the embryo in a high throughput manner. Using our systems, we are capable to (i) improve embryo culture conditions and (ii) diagnose embryos with improved implantation potential.
Label-free microscopy and multi/hyperspectral imaging
Label-free Microscopy: This technique allows for analyzing cells in their native condition, i.e. without being labeled or altered in any way, by means of retrieving cells autofluorescence signals and thus providing essential metabolic information about living tissues. Combined with multispectral methods, we delve deep into the metabolic complexity of embryos and oocytes, revealing insights previously unattainable.
Hyperspectral Imaging: Our groundbreaking approach employs hyperspectral imaging to obtain the metabolic profiles of embryos and oocytes. This methodology allows us to identify key characteristics at the metabolic level, invisible to conventional techniques like brightfield imaging, offering a unique window into fundamental biological processes.
Hardware Techniques: We have extensive expertise in a variety of advanced microscopy techniques, including two-photon microscopy, laser scanning confocal microscopy, spinning disk confocal microscopy and light-sheet microscopy. The integration of these techniques with multi/hyperspectral detection methods enables us to observe biological samples of interest at cellular level with spectral characteristics.
AI and Software Analysis: We employ sophisticated data analysis tools such as spectral histogram analysis and phasor-plot analysis combined with artificial intelligence (AI) methods for classification. These techniques allow us to interpret complex multi/hyperspectral data and draw meaningful conclusions about the viability and quality of embryos and oocytes.
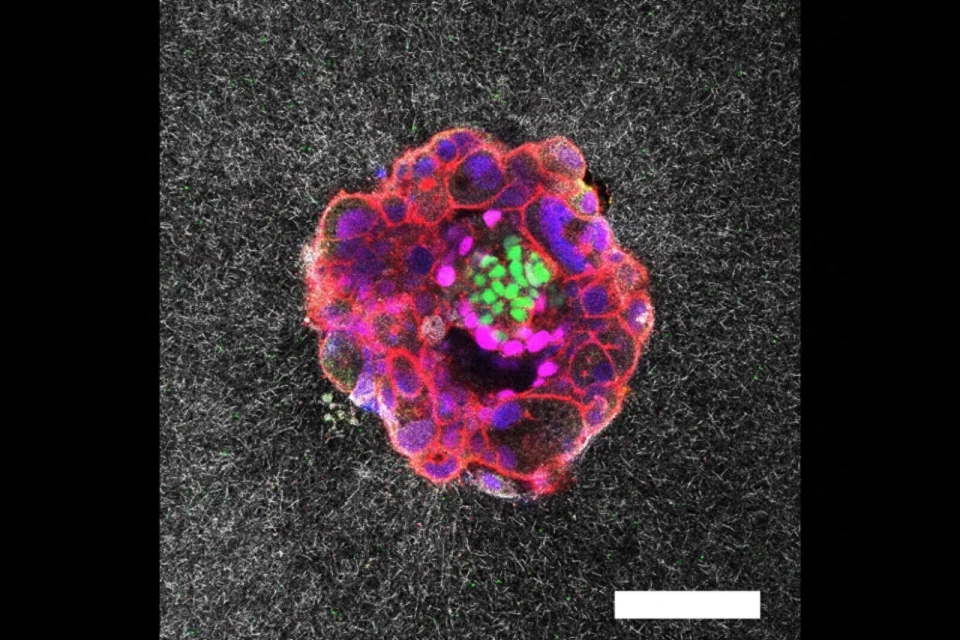
4D (x, y, z, and λ) Hyperspectral Autofluorescent Images of Mouse Oocytes and Blastocysts. The samples were imaged using a Zeiss LSM780 inverted microscope with a C-Apochromat 40x/1.20 W Korr Zeiss objective. The imaging conditions were maintained at 37°C and 5% CO2. A Mai-Tai DeepSee laser provided two-photon excitation at a wavelength λex = 780nm. The emitted autofluorescence spanned from 410 nm to 695 nm and was collected by a 32-channel PMT GaAsP spectral detector. The image captures multiple planes, separated by 2.5μm and 5μm steps for oocytes and embryos respectivelly. The pseudo-color representation of the hyperspectral images was achieved using additive blending to integrate the 32 channel colors for each pixel.
Bioengineering to improve embryo implantation
We have developed proprietary 3D ex vivo hydrogel-based implantation platforms which mimic the uterine microenvironment, allowing the embryo to progress towards post-implantation stages in an amenable way for optical microscopy. Working towards obtaining dynamic control of embryo culture, we have integrated our hydrogels in a microfluidic device allowing for controlled nutrient supply, oxygen concentration and long-term embryo culture. Our 3D ex vivo hydrogel-based implantation platforms allow for drug screening and determination of its impact on embryo implantation and development.
We use our 3D ex vivo implantation platform to understand embryo implantation with a focus on the biomechanics of the system. To this means we quantify the displacement of the matrix generated by the embryos using PIV or DVC algorithms. We look at the forces and resulting patterns embryos are applying in order to penetrate the hydrogel and also how external forces affect embryo implantation.
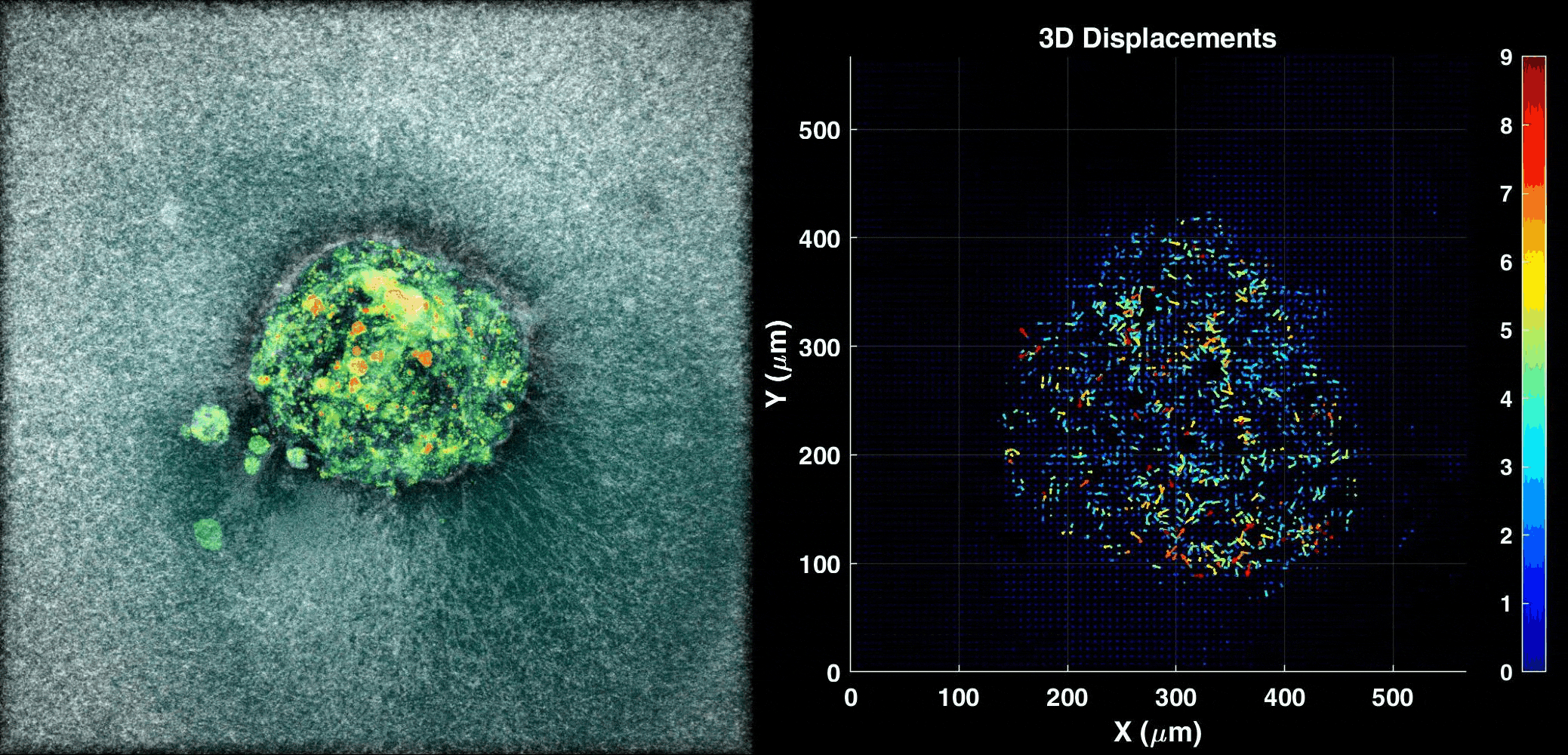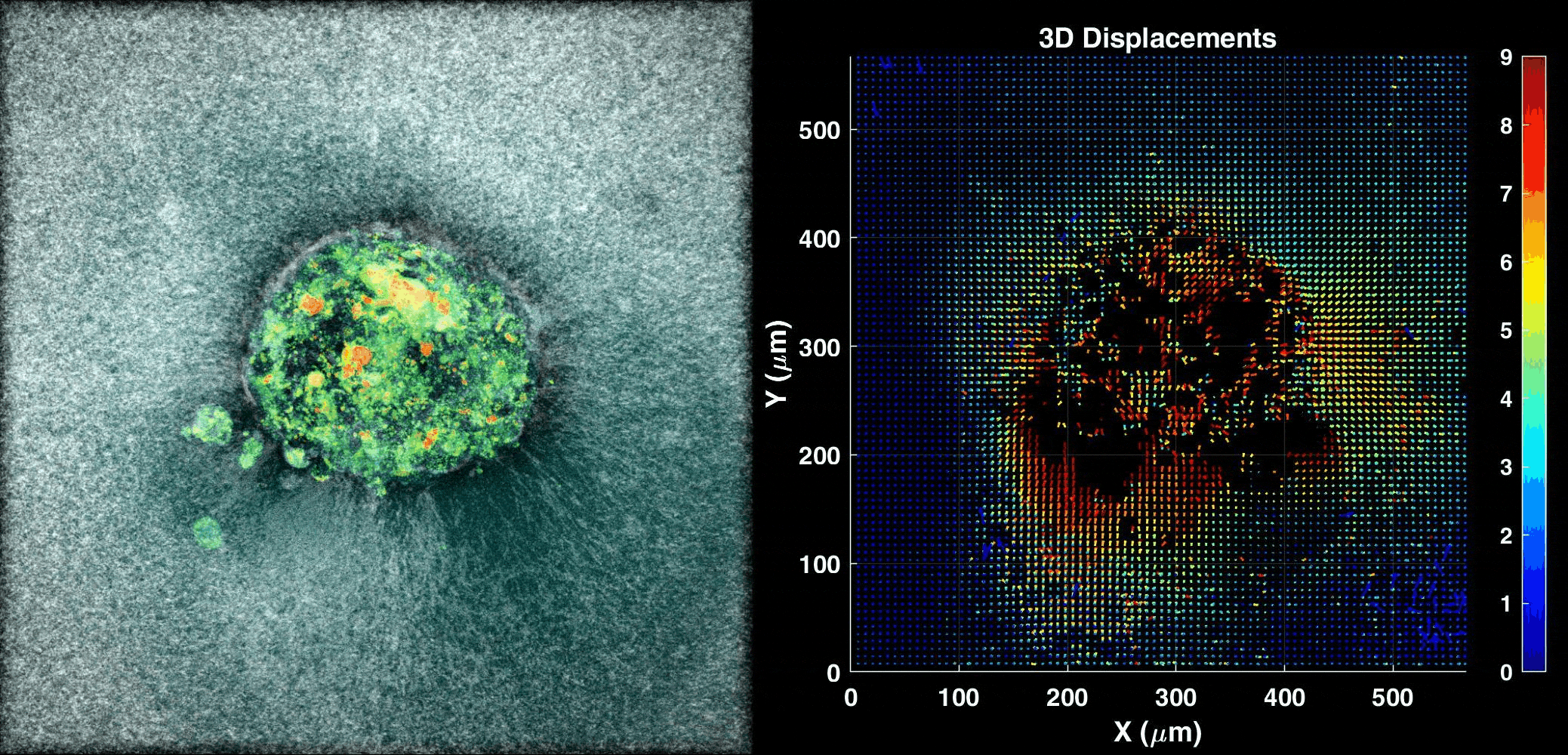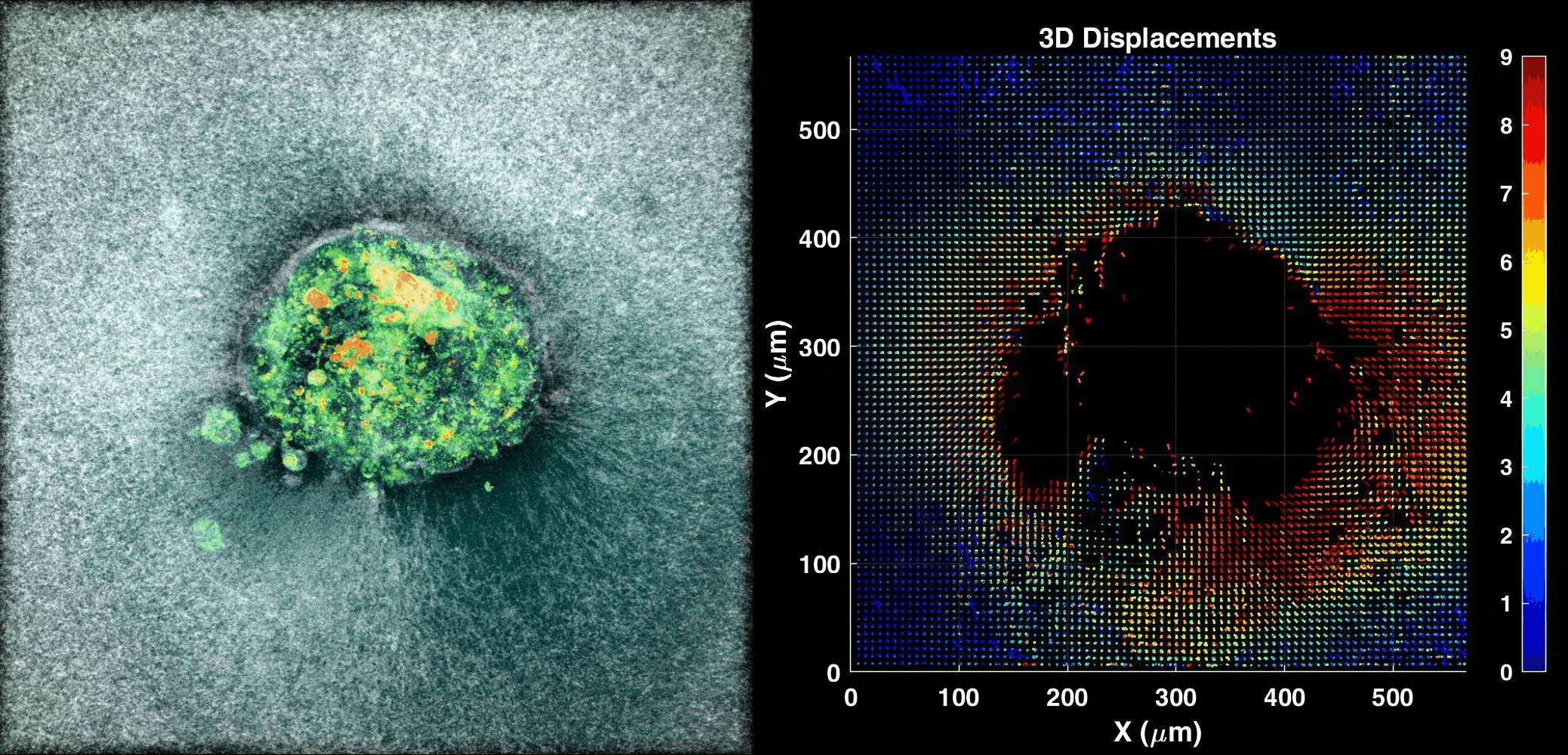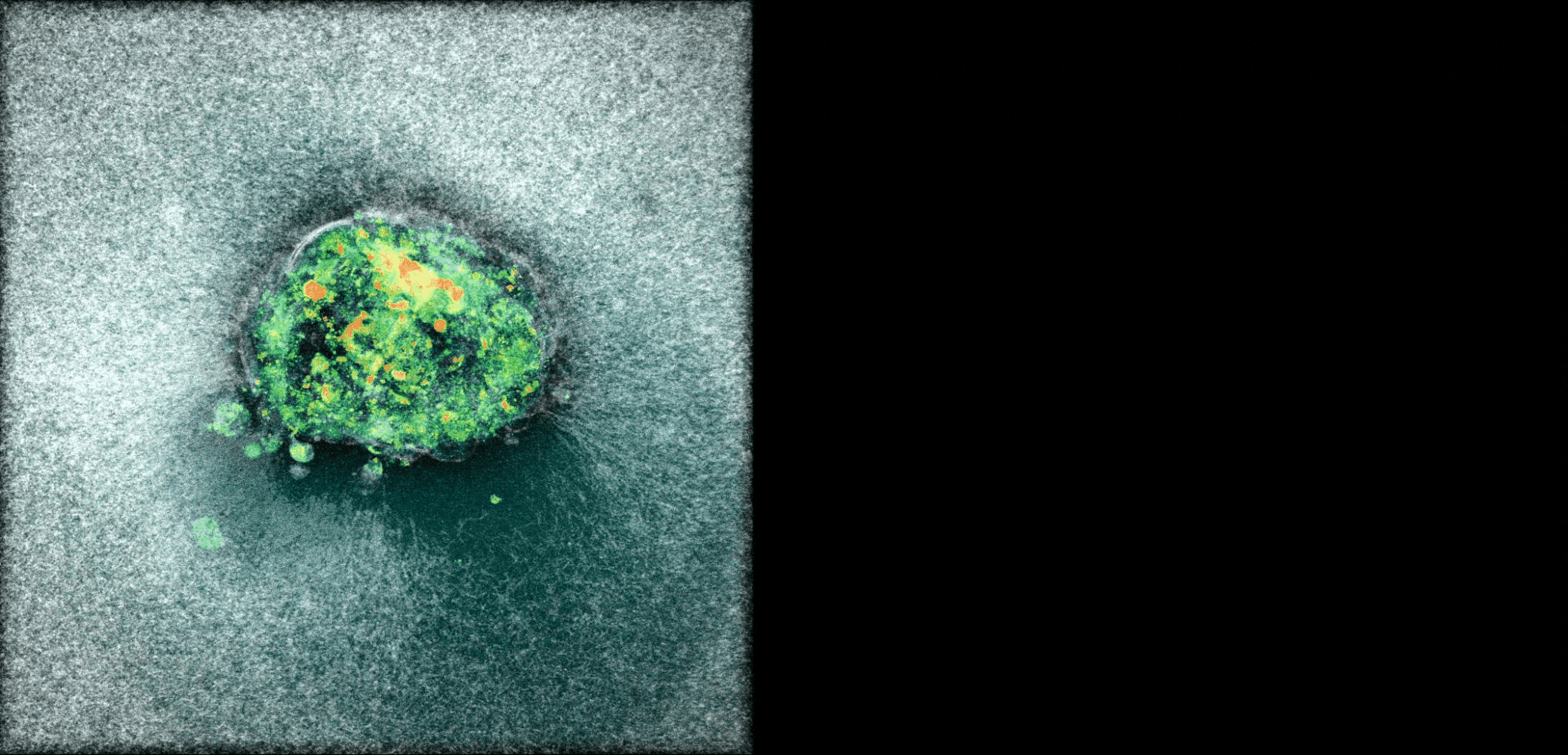
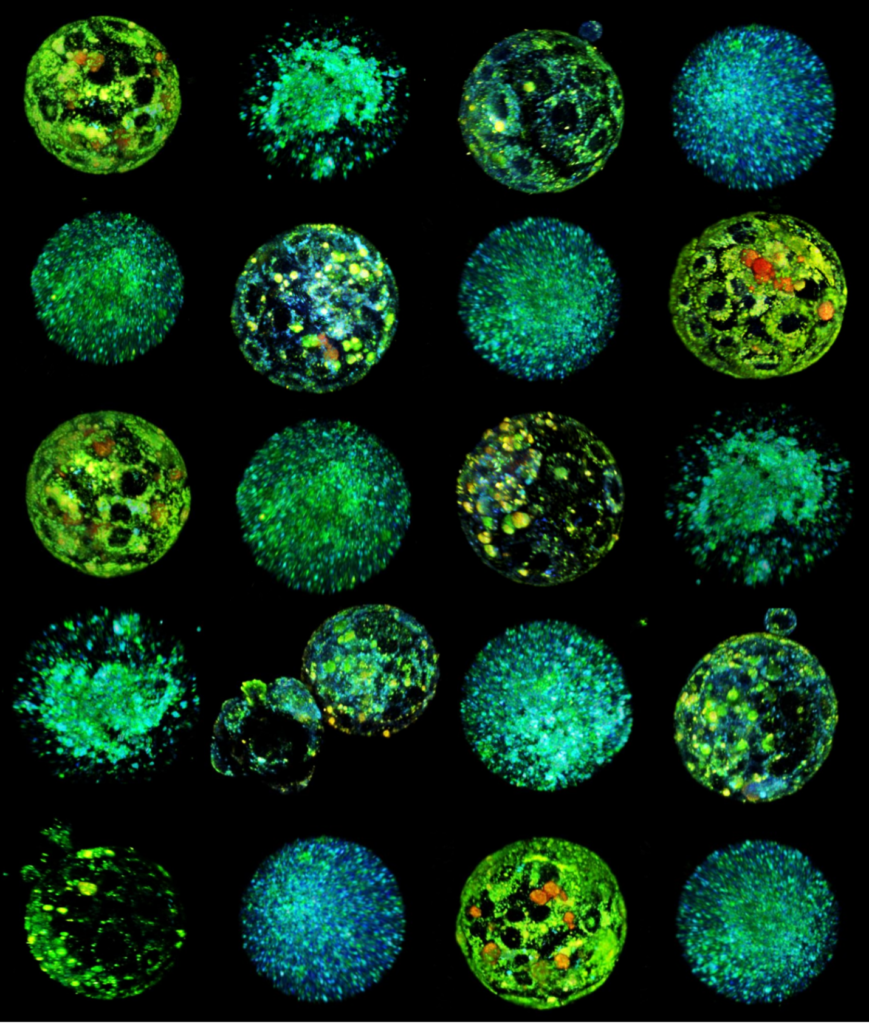
Human Embryo implanting on a 3D platform
Embryo Culture Supplements Development from Human Plasma
Traditionally, embryo culture relied on Human Serum Albumin (HSA) as a key protein component. However, HSA underrepresents the rich composition of proteins present in human plasma. We work with a new-generation of supplements, which go beyond mere albumin, encompassing essential components such as growth factors and globulins, crucial for fostering optimal embryo development. In our group, we test new supplements to enhance embryo development and implantation. Our clinical grade human-derived supplements improve blastulation and implantation rates both in human and mouse embryos, showing superior lineage segregation and spatial organization compared to control counterparts.
The Bioengineering in Reproductive Health is the first Open Innovation Lab research unit at IBEC.
Due to the high translational component of our research, we have established collaboration contracts with the pharma industry, hospitals, and venture capital to bring our technology to the clinics and the market. Our Open Lab is a multidisciplinary environment where embryologists, cell biologists, optical physicists, biophysicists, and business developers synergize to create a unique environment shaped by science and entrepreneurship.
STAFF
PROJECTS
European Projects
| HSMe-ImPredict · Development of non-invasive imaging methodology for improving embryo implantation prediction, via hyper-spectral metabolic profiling (2022-2024) | Marie Curie Individual Fellowship | Samuel Ojosnegros |
National projects
| HYSPLANT · Selección de embriones para fecundación in vitro: predicción del éxito de implantación mediante clasificación metabólica de embriones (2020-2023) | MINECO Retos investigación: Proyectos I+D | Samuel Ojosnegros |
| Prediction of implantation success by hyperspectral metabolic profiling of human embryos obtained by in vitro fertilization (2020-2022) | AGAUR Beatriu de Pinós 2018 | Samuel Ojosnegros |
Private sector
| Estudio del mecanismo de acción de OXO-001 (2020-2022) | Oxolife | Samuel Ojosnegros |
| Evaluación de derivados plasmáticos humanos para el cultivo de embriones (2019-2021) | IVFTECH S.L. | Samuel Ojosnegros |
| Evaluación de derivados plasmáticos humanos para el cultivo de células mesenquimales y CAR-T (2020-2022) | IVFTECH S.L. | Samuel Ojosnegros |
| Prediction of implantation success by single-cell transcriptomic profiling of human embryos obtained by in vitro fertilization (2018-2022) | Scranton Enterprises BV | Samuel Ojosnegros |
Finished projects
| HYSPLANT · Metabolic profiling of in vitro fertilization embryos using hyspectral imaging (2019-2020) | European Commission ATTRACT | Samuel Ojosnegros |
| Embryo on a chip: Smart microdevice development for high-throughput screening embryo implantation (2020) | Tecniospring 2020 | Maria Demestre Viladevall |
NEWS

El IBEC muestra su liderazgo en innovación biomédica en el MWC26 con tres proyectos disruptivos
La semana pasada se celebró en Barcelona el MWC26, el evento internacional de referencia en tecnología y conectividad. El Instituto de Bioingeniería de Cataluña ha estado presente dando a conocer algunas de sus spin-off y tecnologías biomédicas punteras: Nanobots Therapeutics, Lumiris Spectral Solutions y Drug4Sight.

El IBEC consigue financiación competitiva para impulsar las pruebas de validación de 3 proyectos innovadores en salud
El IBEC ha obtenido tres ayudas de la convocatoria de subvenciones del Plan estratégico de investigación e innovación en salud (PERIS). Los proyectos buscan validar una tecnología hiperespectral para mejorar la reproducción asistida, esferoides celulares bioimpresos para combatir la diabetes de tipo I y fármacos activados con luz para restaurar la vista en pacientes con degeneración de la retina.

Bioingeniería para la medicina de precisión en el 18º Simposio del IBEC
El 18º Simposio anual del IBEC se centró en ‘Bioingeniería para la Medicina de Precisión’, una de las áreas clave de aplicación del IBEC. Fueron cerca de 300 las personas asistentes al evento, entre las que se encontraba personal investigador local e internacional. Un ambiente multidisciplinar en el que expertos y expertas de otros centros y la propia comunidad del IBEC tuvieron la oportunidad de presentar sus proyectos e intercambiar conocimiento.

El IBEC y el EMBL Barcelona coorganizan una jornada de colaboración para explorar sinergias
El Instituto de Bioingeniería de Cataluña (IBEC) y el Laboratorio Europeo de Biología Molecular (EMBL) han celebrado hoy una jornada de “matchmaking”. El evento ha reunido a investigadores e investigadoras destacadas de ambos centros con el fin de fomentar la creación de nuevas conexiones y promover el diálogo científico.

El Periódico: El asalto del embrión al útero, al descubierto
Un vídeo hecho en Barcelona documenta por primera vez en un modelo artificial de matriz la implantación, momento crítico y gran desconocido del embarazo humano.
Corriere della Sera: Filmato per la prima volta l’impianto d’embrione umano, il «miracolo della vita» si mostra in tempo reale
Il momento clou della gestazione è stato catturato per la prima volta da alcuni ricercatori dell’Istituto di Bioingegneria della Catalogna. La nuova tecnica potrebbe aiutare gli scienziati a scoprire come prevenire gli aborti spontanei
Metrópoles: Vídeo: registro mostra implantação de embrião humano pela primeira vez
Imagens inéditas revelam como embriões humanos invadem o tecido uterino e explica causa comum de infertilidade

EL MUNDO: Graban por primera vez la implantación de un embrión
Investigadores españoles crean un sistema en el laboratorio que simula las capas externas del útero en 3D. “Ayudará a mejorar la tasa de fecundación in vitro”.

Forbes: Dramatic Footage Captures Human Embryo Implanting In Real Time
Scientists at the Institute for Bioengineering of Catalonia in Barcelona have recorded footage of a key stage of human development in real time, and in 3D, for the first time. In doing so, they witnessed a surprisingly invasive event that they hope could provide insights that may one day help treat infertility.
JOBS
Researcher in Training at the Bioengineering in Reproductive Health Research Group
Ref: RT-SO // Deadline: 24/11/2025
Scientific Liaison at the Bioengineering in Reproductive Health Research Group
Ref: SL-SO // Deadline: 16/06/2025
Postdoctoral Researcher at the Bioengineering in Reproductive Health Research Group/ Unit
Ref: PR_SO // Deadline: 13/11/2024
Embryology Laboratory Technician at the Bioengineering for Reproductive Health Research Group
Ref: PD-SO/Deadline: 04/04/2024
Open postdoc position on embryology at the Bioengineering in Reproductive Health Research Group
Ref: PD_SO/ Deadline: 12/02/2024
Research Assistant at the Bioengineering for Reproductive Health Research Group
Ref: RA_SO/Deadline: 20/11/2023
Postdoc on microscopy at the Bioengineering in Reproductive Health Research Group
Ref: PD_SO//Deadline: 1/11/2023
Postdoctoral researcher at the Bioengineering in Reproductive Health Open Lab (Ref: PD_SO)
Ref: PD_SO // Deadline: 19/01/2023
Embryologist at the Bioengineering in Reproductive Health Research Group (Ref: LT_SO)
Ref: LT_SO // Deadline: 19/01/2023
Biochemist at the Bioengineering in Reproductive Health Research Group
Reference: LT-SO / Until Oct 15th, 2022
PUBLICATIONS
S Ojosnegros, A Seriola, AL Godeau, A Veiga (2021) Embryo implantation in the laboratory: an update on current techniques. Human Reproduction Update, Vol.00, No.0, pp. 1–30.
Martin Plöschner, Denitza Denkova, Simone De Camillis, Minakshi Das, Lindsay M. Parker, Xianlin Zheng, Yiqing Lu, Samuel Ojosnegros, and James A. Piper (2020) Simultaneous super-linear excitation-emission and emission depletion allows imaging of upconversion nanoparticles with higher sub-diffraction resolution. Optics Express 28 (16), 24308-24326.
EQUIPMENT
- Embryo culture laboratory
- IFV workstations in laminar flow hoods
- Microscope
- Micromanipulation-microinjection station
- Embryo biopsy laser
- K-systems incubator
- Cell culture laboratory
- Biosafety cabinets
- Incubators
- Automated cell counter
- Dry warming/thawing system
- Sterile tubing welder
- Tubing sealer
- Centrifuges
- Advanced live imaging: photoconversion, 3D imaging, light scattering, spectroscopy
- Crest spinning disk mounted on a Nikon Ti
- Image analysis workstation
COLLABORATIONS
- Prof. Anna Veiga – Barcelona Stem Cell Bank (IDIBELL) and Dexeus Mujer, Barcelona
- Dr. Montserrat Boada/ Dr. Pere Barri – Dexeus Mujer, Barcelona
- Dr. Ayelet Lesman – Tel Aviv University (TAU), Israel
- Dr. Elena Martínez – IBEC
- Dr. Francesco Cutrale, University of Southern California (USC), USA
- Dr. Manuel Irimia – CRG, Barcelona
- Dr. Javier Ramón – IBEC
ENTREPRENEURSHIP
· Jorge Fuentes,
Business Strategy, A_Ventures, Barcelona, Spain
 |  |  | |
 |  |
Follow us on Twitter: @Biorephealth