About
Cells continuously sense their environment by integrating multiple input signals from neighbouring cells and respond to them with a fast-flowing cascade of abundant outputs. In turn, these outputs modify the environment.
Taking a set of distinct cell types, this spatiotemporal combinatorial phenomenon can presumedly lead to a vast number of multicellular spatial patterns (Figure 1). Still, only a relatively limited number of stable multicellular spatial patterns with functions (and dysfunctions) emerge from all possible coordination events amongst multiple cell types. For these patterns to repeatedly arise in distinct organisms, a set of principles driving the spatial organization of multiple cell types must be conserved; however, these principles remain largely unknown as we have only recently begun to understand how multicellular organization works. Being able to read, anticipate the outputs, and eventually control the principles behind the spatial organization of multiple cell types are, to us, some of the most fascinating problems of modern biotechnology. Advances in this area are expected to be key to manipulate tissue function and, therefore, they will undoubtedly bring numerous benefits to society: from a deeper understanding of the natural world to better cell-based therapies, diagnostics tools, and engineered tissues for precision and regenerative medicine.
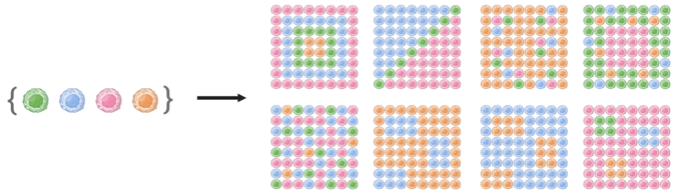
Figure 1. A set of cell types can theoretically arrange in a vast number of spatial patterns.
In the Spatial Biotechnology group, we are making headway on this problem via the in-depth study of clonal behaviors in solid tumors. The number of cancer patients is expected to skyrocket in the next few decades in part due to the progressive aging of the population. Therefore, it is imperative to improve and expedite cancer management. A common reason for the failure of current therapies in solid tumors is that they are highly heterogeneous entities, and a major source fueling this heterogeneity is clonal diversity, which encompasses genetic variability and epigenetically regulated cell states. Clonal diversity significantly influences tumor growth, metastasis, and therapy response. It is increasingly clear that the spatial organization of cancer cell clones is a crucial factor in solid tumor development, as cell fate is intimately linked to the local environment. However, our understanding of the rules governing clonal spatial organization in tumors is limited, largely due to the absence of mature experimental and computational frameworks to investigate clonal behaviors in the native tissue context.
The goal of the Spatial Biotechnology group is to develop high-throughput and spatial perturbation technologies to understand how cellular clones spatially organize to drive solid tumor development, with the long-term goal of designing better cancer therapies and tools to predict clinical outcomes.
To achieve this goal, our group develops experimental and computational approaches for spatial multi-omics, and it applies them to interrogate and engineer a variety of clonal behaviors in solid tumors from clinical sources, murine models, and in vitro tissues. This interdisciplinary work builds on our expertise in developing highly multiplexed tissue imaging technologies, super-resolution, mass spectrometry, and multimodal microscopy, pooled genetic screens, assay automation strategies, immune cell engineering approaches, and computational tools for deconstructing molecular and cellular spatial patterns in cells and tissues (Figure 2).
In the Spatial Biotechnology group, we focus on some of the most pressing questions in the field, specifically:
1) How is clonal cooperation established and maintained?
The widespread clonal heterogeneity of human tumors implies that several ecological interactions are in play, together with clonal competition. Indeed, recent evidence supports that tumor evolution could be also driven by clonal cooperation, where growth equilibrium of two or more clones might be a path to optimal fitness. Using genome editing tools, pooled screens, and super-resolution multiplexed imaging, we interrogate molecular signatures of tens of genetically engineered cancer cell clones in the native tissue context in in vivo murine models. This work aims to characterize molecular determinants promoting clonal cooperation in cancer, which would provide causal understanding on how genetically unique clones arrange within a tumor and regulate tumor growth.
2) How does metabolite accessibility regulate the growth of neighboring cancer cell clones?
Distinct environmental conditions simultaneously arise in multiple regions of a tumor providing selective pressure to the variable cancer cell population at place, further increasing heterogeneity or dramatically shifting the composition of the clonal pool. For instance, metabolic reprograming of cancer cells can result in localized and differential enrichment of metabolites. Using automation strategies and highly multiplexed imaging, we characterize clonal cancer cell behaviors in hundreds of in vitro 3D tumors growing in metabolically defined environmental conditions. This work aims to identify clonal signatures in specific metabolic environments, which would provide clues on how the environment regulates clonal growth and potentially inform future therapeutic interventions.
3) How do clonal populations and microenvironments evolve in primary tumors?
The tumor microenvironment, which includes resident immune and stromal cells as well as the extracellular matrix, is closely linked to the evolution of the cancer cell population. Using spatial transcriptomics and histopathology-grade proteomics, we correlate molecular features that are predictive of clonal growth in a variety of patient cohorts in collaboration with clinicians. This work aims to identify clonal patterns in clinical tumor samples, which would inform about spatial features correlating with clinical outcome for better disease management.
4) How could adoptive T-cell transfer therapies be improved for solid tumor treatment?
Some of the major obstacles to increase the success of adoptive T-cell transfer therapies for solid tumors is limited T-cell infiltration, persistence, and functionality. Using immune cell engineering approaches and highly multiplexed imaging, we track the tumor location of adoptive T-cell transfer therapies and assess their functional status in the native tissue environment of models permissive to T-cell infiltration. This work aims to identify molecular determinants of productive T-cell fitness, which would provide alternative options to improve current cell-based therapies for cancer.
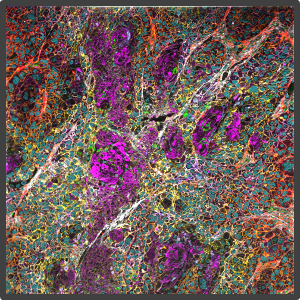
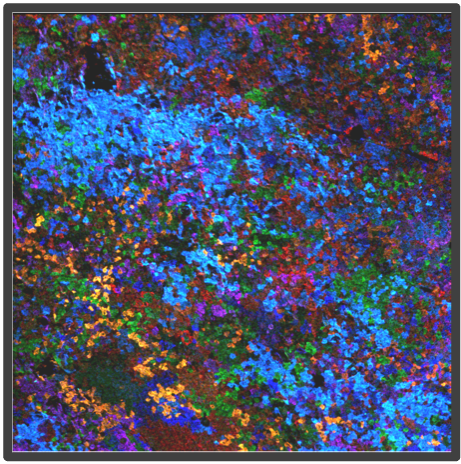
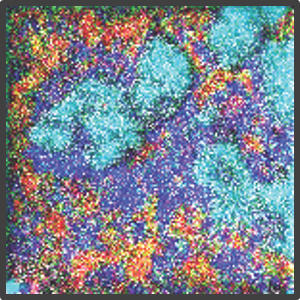
Figure 2. Representative highly multiplex images in a variety of models analyzed by Multiplex Ion Beam Imaging (MIBI), a spatial proteomics technology that detects 40+ antibodies in tissue sections using isotopes. (Left) Histopathology-grade MIBI in a section of human squamous cell carcinoma. (Middle) Clonal tracing studies using MIBI in a tumor grown in vivo. (Right) Super-resolution multiplex imaging of the nucleus of a cisplatin-treated TYK-nu cell, an ovarian cancer cell line.
Staff
Projects
Publications
Collaborations
- Prof. Garry Nolan
Department of Pathology, Stanford University, United States - Prof. Julien Sage
Department of Pediatrics, Stanford University, United States - Dr. Alexandros Drainas
BioMed X Institute, Heidelberg, Germany - Dr. David McIlwain
University of Nevada, Reno, United States - Dr. Sizun Jiang
Center for Virology and Vaccine Research, Harvard Medical School, United States - Dr. Franzi Blaeschke
German Cancer Research Center (DKFZ), Heidelberg, Germany - Dr. John Hickey
Department of Biomedical Engineering, Duke University, United States - Dr. Jolanda Sarno
Tettamanti Research Center, University of Milano-Bicocca, Italy - Prof. Miriam Cuatrecasas
IDIBAPS, Barcelona - Dr. Jordi Camps
IDIBAPS, Barcelona
News

El IBEC se refuerza con tres nuevos grupos de investigación en terapias avanzadas y emergentes
El IBEC estrena el 2024 con la incorporación de tres nuevos grupos de investigación que serán liderados por Manuel Salmerón Sánchez, Zaida Álvarez Pinto y Xavier Rovira Clavé. Con estas incorporaciones, el IBEC fortalece su posicionamiento en el campo de las terapias avanzadas y emergentes.

El ERC apuesta por el proyecto “SpaceClones” para comprender la evolución clonal de los tumores
Xavier Rovira Clavé, actualmente investigador en la Facultad de Medicina de Stanford, ha sido seleccionado en la convocatoria “Starting Grant” del Consejo Europeo de Investigación (ERC) para llevar a cabo su … Read more
Jobs
Predoctoral Researcher at the Spatial Biotechnology Research Group
Ref: PhD_XR//Deadline: 22/03/2024


 ibecbarcelona.eu
ibecbarcelona.eu