
Former Members
Dr. Jérôme Noailly | Principal Investigator
Now: SIMBIOSYS (Simulation, Imaging and Modelling for Biomedical Systems), UPF
Dr. Damien Lacroix | Group Leader
Now: University of Sheffield
About
Biomechanics (spine, lower limbs); Mutiphysics (cartilage, intervertebral disc, artery); Biophysics (intervertebral disc nutrition, cytokines); Computational analyses (finite element element, numerical optimisations, stochastic modelling); In vitro experiments (dynamic culture, tissue/biomaterial mechanical characterisation)
Research in the group of Biomechanics and Mechanobiology focuses mainly on (i) the interactions between tissue multiphysics and biological processes, and (ii) how these interactions can affect the functional biomechanics of organs. Numerical methods based mostly but not exclusively on FE modelling are used to describe both the tissues at the organ level, and the tissue-cell interactions at the tissue and cellular levels. The numerical concepts developed are tested against in vivo and in vitro data, which allows model validations. Emphasis is given in the study of load transfer of organ conditions onto the cells or onto tissues, with or without treatment simulations. Calculations are based on mechano-regulation and/or on biophysical concepts to predict different cell environments over time.
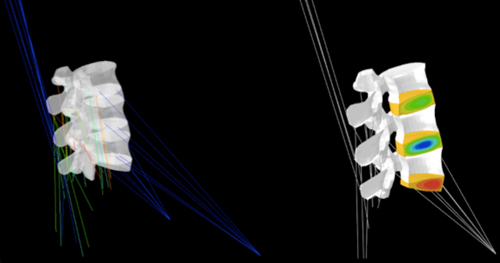
Most tissue and biophysical models developed so far aimed to study one of the most complex organs of the musculoskeletal system, namely the spine. Thorough knowledge about the functional biomechanics of the lumbar spine has been acquired along the time in relation to computational simulations (J Biomech, 40, 2414-25; Biomech Model Mechanobiol, 10, 203-19). In order to capture as best as possible the communications between organ and tissue biomechanics, studies of advanced tissue models have been performed, in relation to the vertebrae (Mater Lett, 78, 154-58), to the intervertebral discs (J Mech Behav Biomed Mater, 4, 124-41; Comput Meth Biomech Biomed Engin, 16, 923-8) and to the muscles (J Biomech, 45, S484).
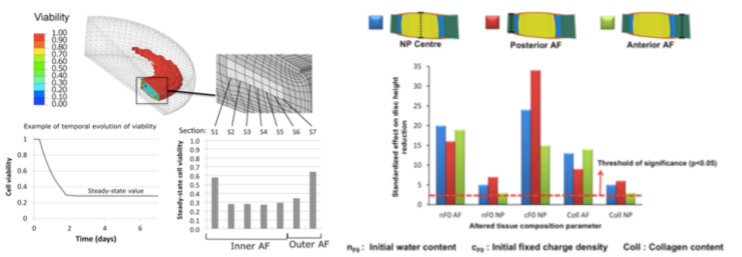
In particular, these models allowed thorough identification of the tissue parameters expected to alter cell nutrition in a deforming intervertebral disc (PLoS Comput Biol, 7, e1002112), leading to further relations between tissue condition and cell viability (Poromechanics V, 2193-2201). Care is also taken to assess the physical meaning of the tissue model parameters, and the mechanistic aspect of the simulation work is supported by both stochastic modelling and bioreactor experiments.
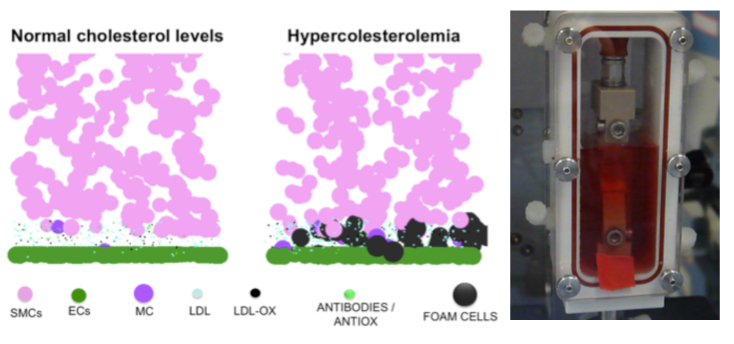
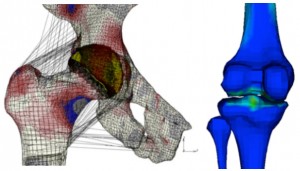
The numerical stability of these models is also one target of the explorations performed within the group (J Mech Behav Biomed Mater, 26, 1-10), in order to ensure the coupling to lower scale biophysical models. Also, models have been used to for implant simulations focussed either on clinical (J Appl Biomat Biomech, 4, 135-42), or on design questions (Eur Spine J, 21, S675-87). Beyond the spine domain, both knowledge and know-how acquired are being transferred to the exploration of the cardiovascular system. Also, on-going clinical collaborations are contributing to the adaptation of the numerical methods to study problems and treatment solutions related to the lower limbs (J Biomech, 45, S163).
Projects
EU-funded projects
| My Spine: Functional prognosis simulation of patient-specfic spinal treatment for clinical use | EU – Cooperation – FP7-ICT | Jérôme Noailly |

