About
In the group we advance cross-disciplinary research at the interface between biology, physics and engineering by studying the mechanical biology and the biological mechanics of pathological development and disease progression. Specifically, we focus on soft tissue morphogenesis – the process by which a tissue takes or lose shape.
Controlling the biological and mechanical behaviour of cells at the molecular level is a central axiom of bioengineering in its pursuit of tissue and organ regeneration. However, steering living-cell aggregates towards predesigned tissue architectures requires the concertation of factors such as the signalling, shape, force, adhesion and motility of single cells at length- and time-scales still largely unknown. Moreover, increasing evidence points out that the emergence of collective behaviour in cellular assemblies such as tissues is governed by mesoscale physical principles that may also instruct cell biological function in an independent manner. Yet these principles at the tissue scale cannot be predicted from biochemical principles at the single-cell scale.
Our research aims at understanding the role of cell mechanics in tissue (mal)formation by harnessing the mesoscale mechanical strategies that cellular collectives adopt to determine tissue form and function in physiological and pathological conditions in vivo and in vitro.
By so doing, we wish to provide bioengineers with a modern swatch of fundamental principles that can be utilised to master synthetic morphogenesis and tissue design for regenerative and therapeutic purposes.
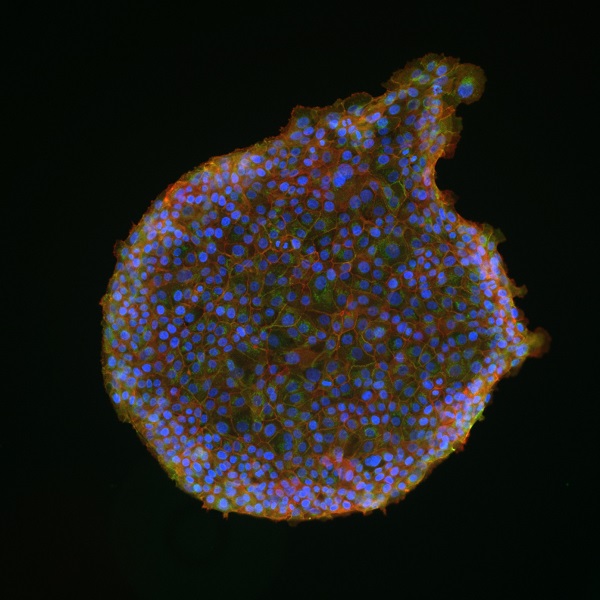
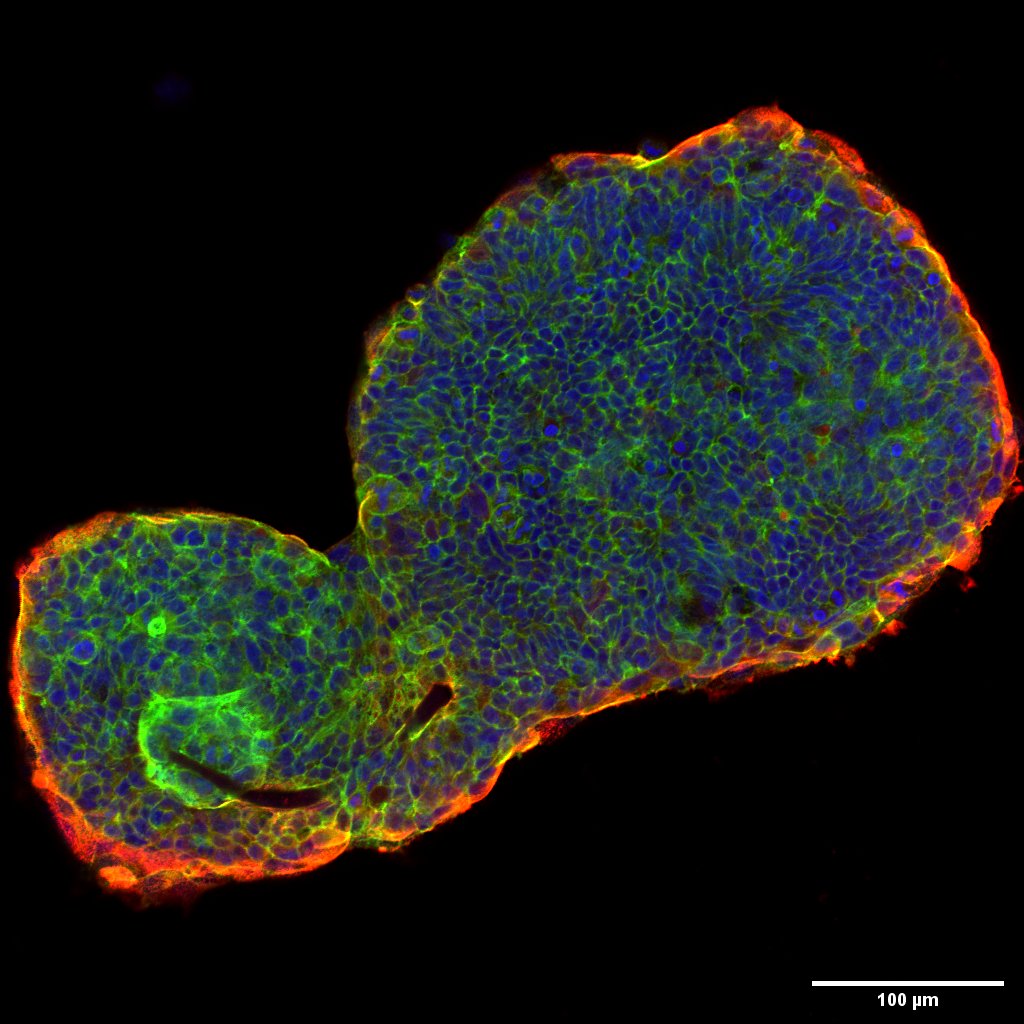
Projects
| FINISHED PROJECTS | FINANCER | PI |
|---|---|---|
| CancerMechReg Regulacion biomecanica de la progresion del cancer (2016-2019) | MINECO, Proyectos I+D Excelencia  | Vito Conte |

